
David G. King
Annotated list of publications and presentations
Each of the entries below is followed by a brief personal comment.
Reading from the bottom up, this list charts the trajectory of my research career, from graduate school through retirement.
The comments also map the origin and subsequent history of the "evolutionary tuning knobs" metaphor.
before 1981 | 1981-1990 |
1991-2000 | 2001-2010 |
2011-2020 | since 2020 | Epilogue
My career has been ultimately inspired by my interest in natural history. Since childhood,
this interest has stimulated a desire to understand patterns of structure and genetics underlying adaptation, including
the marvellous evolutionary malleability of animal behavior (cf. King 2013).
I have grown ever more impressed by "extremely minute parts so shaped and situated as to form a marvelous organ" (to quote
17th-century microscopist Marcello Malpighi), especially by the exquisitely organized
complexity of the nervous system (whose marvelous cells Santiago Ramón y Cajal
memorably called "mysterious butterflies of the soul.") In each generation, such complexity somehow emerges anew from a single
fertilized egg.
Most of the peer-reviewed publications listed below are available somewhere on the internet,
at least in abstract; full-text PDFs may be requested by email.
Meeting presentations (posters and slide shows; not peer-reviewed) are by nature ephemeral (except for the pdfs archived here).

(2025b) King, D.G. Evolvability
and lineage selection. Science E-Letter (published 19 March 2025).
This contribution is an e-letter (not appearing in print) commenting on a research article by Barnett, et al.,
Science 21 February 2025, Experimental evolution of
evolvability. It appears farther down on the same webpage as Barnett et al.'s article. From
my point of view, the principal virtue of this piece is that it appears in a fairly prominent venue and
cites my 2024a and 2024b publications (below). (My publishing career might conclude
with this short note.)

(2025a)
King, D.G., Tandem repeat polymorphisms shape local adaptation. Trends in Ecology
& Evolution 40: 331-334, https://doi.org/10.1016/j.tree.2025.02.004.
This essay highlights a report exploring "the extent to which STRs, especially those that that are linked to adaptive traits, could be shaped by the environment" [Ranathunge and Welch (2024) Clinal Variation in Short Tandem Repeats Linked to Gene Expression in Sunflower (Helianthus annuus L.), Biomolecules 14:944 https://doi.org/10.3390/biom14080944]. The authors have kindly shared with me that their report, the culmination of a multi-year project including several related articles, was inspired in part by two 1997 articles by Kashi, Soller, and myself.
My research career began with ecology, with participation in an NSF-sponsored program while I was still in high school.
So I was delighted to finally land a paper in Trends in Ecology & Evolution. My proposal for this paper was my fourth attempt to publish in the journal;
previous attempts were eventually accepted into Endeavour (1997) and Trends in Genetics (2006, 2024).
(2024b) King, D.G.,
Evolving a favorable distribution for mutation effects. Trends in Genetics
40:819-821. doi: 10.1016/j.tig.2024.07.009.
[email for PDF: dgking@siu.edu].
This essay was designed to call attention to my just-published article in The Journal of Physiology (below).
It is a much shorter, more readily digestible version of that article, written for a venue where it is more likely to be noticed by geneticists.
Hopefully, it might also encourage someone to explore the potential of indirect selection through computational modelling of population genetics.
I wish I could have offered a citation for Ranathunge and Welch's 2024 report in Biomolecules,
but their paper appeared too late for inclusion.
(While preparing this ms., I came across a paper that I had not previously noticed, presumably because it does not cite any of the reviews that had become familiar to me:
"Engineering in Genomics: variable number tandem repeats as agents of functional regulation in the genome," G. Breen et al. [2008], Engineering in Medicine and Biology
Magazine 27: 103-104, 108. This reminds me that different communities of biologists can sometimes remain fairly oblivious to one another.)
(2024a) King, D.G.,
Mutation protocols share with sexual reproduction the physiological
role of producing genetic variation within 'constraints that deconstrain'. The Journal of Physiology 602: 2615-2626; doi.org/10.1113/JP285478.
[abstract] [free-access pdf]
Unexpectedly, after nine years of retirement, I stumbled into an invitation to write
for a special "physiology of evolution" issue of the venerable Journal of Physiology. This
essay reviews material explored
on several previous occasions (listed below, especially posters in 2014 and 2015, also
abstracts from 1985 and 2010), but this time written for J. Physiology readership.
Some readers might find it incredible that the outdated views of mutation addressed in this essay might still need refutation.
Yet even in 2023, in Nature Reviews Genetics 24:3, we can read that
"Sturtevant's [1937] views on the evolution of mutation rates are likely to continue to influence future efforts to understand what remains
a fundamental question in biology." My primary impetus for this ms. was to continue challenging just such misleading but persistent influence.

(2015) King, D.G., Might constrained mutability
be as advantageous as sex? (In this pdf, the poster display follows the "Further reading" list.)
This poster, like the two immediately below, takes direct aim at the classic but misleadingly simplistic
assertion, famously declared by George Williams in 1966, that "[N]atural selection of mutation rates has only
one possible direction, that of reducing the frequency of mutation to zero... So evolution takes place, not so
much because of natural selection, but to a large degree in spite of it."
For several years, I thought that this post-retirement meeting presentation would represent my
final effort to advocate for the idea of mutation protocols
as metaptation. This was somewhat disappointing, since posters are ephemeral
(except as archived here). However, in 2023 I was invited to contribute an article to a special
issue on "physiology of evolution" in The Journal of Physiology.
(2014) King, D.G., A lesson from sex:
Abundant variation can be worth its high price. (In this pdf, the poster follows an abstract and reading
list.)
This poster (presented for the Society for Molecular Biology and Evolution annual
meeting in San Juan, Puerto Rico), like the ones immediately above and below, challenges the misguided presumption
that, in Sturtevant's oft-quoted words from 1937, "the vast majority of mutations are unfavorable... [M]utations are
accidents, and accidents will happen." But this presumption ignores the simple fact that sexual reproduction
is, fundamentally, a vast source of potentially advantageous mutations: Practically every instance of meiotic
recombination creates DNA sequences that have never previously existed. And nearly all of these are NOT unfavorable.
☞ Simply because they are seldom deleterious, the products of reciprocal
recombination are often explicitly excluded by definition from textbook descriptions of mutation. For example,
in the glossary of one highly regarded textbook (Douglas Futuyma, Evolutionary Biology 3rd ed., 1998), "mutation" is defined
as any alteration of the genome "that is not manifested as reciprocal recombination [emphasis added]."
(2013b) King, D.G., Whence variation?
Rethinking mutation and evolvability. (In this pdf, an abstract and a reading list follow the poster.)
This poster, together with the two immediately above, takes direct aim at the classic but misguided
dogma, reiterated as recently as 2012 by Elmore et al. (Genes Genomes
Genetics 2: 1643-1649), that "a well established and supported tenet of
evolutionary theory is that, because most new mutations are deleterious, selection in all organisms will act
to reduce mutation rate toward the physiology- or selection-imposed minimum."
(2013a) King, D.G. What
can giant axons tell us about genetics and evolution? International
Society for Neuroethology Newsletter, July/August 2013, pp. 5-7.
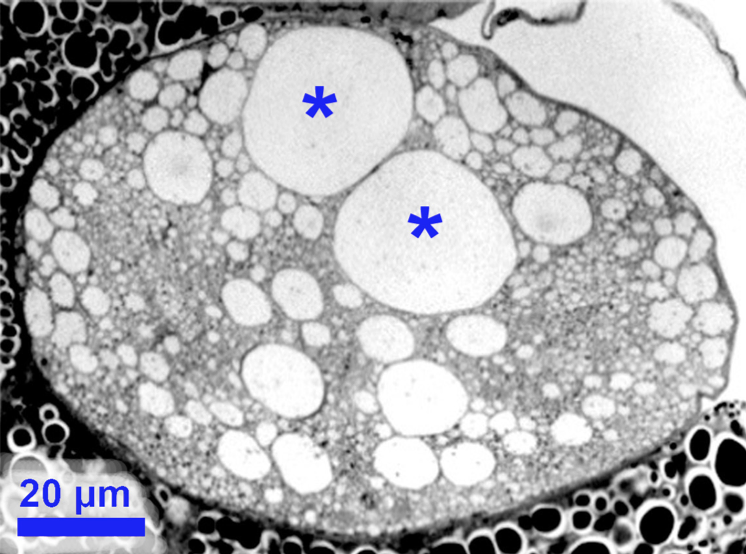 |
A pair of giant axons in a small fly
(note additional large-axon pairs). |
As my retirement approached, I prepared this essay -- illustrating distinctive patterns of nerve fibers in several
species of flies -- as a "swan song" to my interest in specialized neuronal form and function. This essay encapsulates the logic which
has guided my career for several decades.
☞ The idea of individually specialized nerve cells has fascinated me since 1970, when I was introduced to it
during my first "rotation" in graduate school in Ted Bullock's lab at UCSD.
There I was assigned to record electrical activity from one particular cell (R-15, "the parabolic burster") in the
abdominal ganglion of Aplysia.
My doctoral dissertation
(1976b and 1976c, below) focused on several individually-identifiable cells in the
stomatogastric ganglion of the California spiny lobster.
My postdoctoral work was directed toward understanding unique giant axons in Drosophila.
During the early 1980s, I began examining a variety of different fly species in hopes of pinning down the phylogenetic origin
of those curious giant axons. (Although a simple story of giant fiber evolution continues to elude me, the specimens collected for
that purpose served admirably for my 1991 report on the evolutionary origin of the dipteran cardia.)
Hoping to stimulate more widespread interest in the evolution of individually-identifiable nerve cells, I reported further details
of fly neurons at several annual meetings of the Society for Neuroscience. When those presentations received very little attention,
I put neurobiology aside in favor of my transition to studies of evolutionary genetics. Yet even this transition was indirectly
driven by my desire to understand how individually-identifiable nerve cells could evolve, particularly when the totality of
information in a genome would seem inadequate (at least to me) to specify such precision circuitry.
This essay expresses my conviction that the goal of understanding nervous system organization
might benefit by addressing evolutionary processes. (For a related, extended discussion, see my 2001 online-only
essay, "Evolution of neural circuits." Also cf., 1988b, 2007c.)
| What the devil determines each particular
variation? What makes a tuft of feathers come on a cock's head, or moss on a moss-rose? |
 |
| Charles Darwin
(letter to T.H. Huxley, 1859) |
|
(2012)
King, D.G. Indirect Selection of
Implicit Mutation Protocols. Annals of the New York Academy of
Science 1267:45-52.
doi: 10.1111/j.1749-6632.2012.06615.x
[author preprint] [email for PDF: dgking@siu.edu]
At the time, this essay felt like the culmination of my work on the implications of indirect selection for evolutionary processes.
Subsequent poster presentations, at meetings held in 2013, 2014,
2015, and also listed above, have offered some further elaboration. A dozen years later
I found opportunities for more forceful presentations, published in 2024.
(2011d) King, D.G. Indirect
selection of local mutation rates (abstract only). DIMACS Conference on Effects of Genome Structure and Sequence on the Generation of
Variation and Evolution, August 9 - 11, 2011.
In 2011 I was invited to participate in a conference at the Rutgers University Center
for Discrete Mathematics and Theoretical Computer Science. My abstract, initially published in proceedings of this
conference, was subsequently expanded into an essay (2012, above), published in a collection edited by
Lynn H. Caporale with the
same title as the conference, "Effects of Genome Structure and Sequence on the Generation of Variation and Evolution," in the Annals
of the New York Academy of Sciences, Volume 1267 (2012).

(2011c) King, D.G. Genetic
Variation Among Developing Brain Cells. Science E-Letter (published
16 May 2011).
This letter-to-the-editor (continuing our campaign to publicize the evolutionary role of repetitive DNA) was submitted
in response to a news article by Gretchen Vogel (
Science 332:300-301, 2011), "Do Jumping Genes Spawn Diversity?" (This is an e-letter, not appearing in print;
it appears farther down on the same webpage as Vogel's article.)
(2011b) King, D.G. Light on genetic dark matter (letter to the
editor). Science News 179(7):30 (26 March 2011).
Another letter-to-the-editor in our campaign to publicize the evolutionary role of repetitive DNA, this time in a
science magazine published for the educated general public.
(2011a) King, D.G. Evolution of simple sequence repeats as mutable
sites. In: Anthony Hannan, ed., Tandem Repeat Polymorphisms: Genetic
Plasticity, Neural Diversity and Disease, Landes Bioscience and Springer
Science+Business Media. Adv Exp Med Biol. (2012) 769:10-25.
[preprint] [email for PDF: dgking@siu.edu]
Unconstrained by any publisher-imposed word limit, in this article I was able to develop a more thorough consideration
of simple sequence repeat evolution than I have accomplished anywhere else, before or since (cf. 2012).
Here, in addition to reviewing both classical and more-nuanced discussions of natural selection for mutation-rate,
I consider the significance of the phylogenetic distribution of tandem-repeat sites, many of which are conserved across vast spans of
evolutionary time. Such conservation comprises evidence, which I find especially
compelling, that the evolution of tandem repeats has indeed been driven by more than mutation pressure and genetic drift. Positive
(albeit indirect) selection -- favoring, preserving, and shaping the mutagenic potential of tandem repeats -- has surely been involved
as well.
(Due to citations added in proof, the author preprint available here differs slightly from the print
version of this article.)
 |
Image by Milo Winter,
Aesop's Fables, 1919
(adapted
from Wikipedia Commons) |
(2010b) King, D.G. Metaptation: Metaphors
for genome evolution. In:
Structural and Functional Diversity of the Eukaryotic Genome, Verhandlungen
des naturforschenden Vereines in Brünn, Specialausgabe, p. 40.
This abstract, which offers an updated definition
for the word "metaptation" (first introduced in 1985), was written for an international workshop held
at the Mendel Museum in the former
St. Thomas Abbey (Brno, Czech Republic) where Gregor Mendel grew his peas. It might be my only publication to include
an exclamation point!
My invitation to speak at this
workshop came from Ed Trifonov, who had been coauthor on
our 2006a article in The Implicit Genome. The image at right links to the slideshow for
my presentation. Most of the other presentations at this meeting were dense with molecular information that went way
over my head. Cheeky of me, a neuroscientist, to speak at a molecular genetics seminar in Mendel's abbey.
I took advantage of my post-retirement 2024 Journal of Physiology paper
to slip the word "metaptation" into mainstream biology literature.

(2010a) King, D.G. A hypertext
resource for self-directed learning. (poster pdf)
Throughout my career, my principal teaching assignment has been histology. In 2000 I began building a website
emulating my role as instructor, to help students learn that subject. This poster describes the resulting resource.
The website itself, consisting of several hundred webpages of extensively hyperlinked
text and images, was initially posted at a time when internet resources were not as extensive as they have since become.
By 2010 this website had acquired sufficient popularity that a Google search for the single word "histology" returned this site in first
place out of several million hits. This website remains a work in progress. For obscure reasons it was deleted from
the SIU School of Medicine webserver in 2021; nevertheless, after being resurrected at its current URL,
histology.siu.edu, this website has regained its visibility to Google searching. I have been extremely gratified to receive "fan mail" from
around the world, expressing appreciation for the quality of this site.
(2009b) King, D.G., Why do so many
neurodevelopmental genes contain repeat-based mutable sites? (poster pdf)
I prepared this poster for the 2009 meeting of the Society for Neuroscience (the last such meeting which I have attended),
hoping to call some attention to especially intriguing observations of tandem-repeat conservation
in two papers published that same year.

(2009a) King, D.G., and Y.
Kashi Heretical DNA sequences? Science 326: 229-230.
This letter-to-the-editor was written to challenge simplistic application of the "conservation equals function" paradigm in evolutionary
genetics, while publicizing the intriguing results recently published by Riley & Krieger.
(cf., 2009 poster).
(2008b) King, D.G., Facilitating Evolution: Adaptation and Simple Sequence Repeat Polymorphism. (poster pdf)
This poster was prepared for an ecology symposium; an abstract and a reading list follow the poster.
 (2008a)
Fondon III, J.W., E.A.D. Hammock, A.J. Hannan, and D.G. King Simple sequence repeats: Genetic modulators
of brain function and behavior. Trends
in Neurosciences 31: 328-334. doi: 10.1016/j.tins.2008.03.006.
[authors' prepublication MSWord
document] [email for PDF: dgking@siu.edu]
(2008a)
Fondon III, J.W., E.A.D. Hammock, A.J. Hannan, and D.G. King Simple sequence repeats: Genetic modulators
of brain function and behavior. Trends
in Neurosciences 31: 328-334. doi: 10.1016/j.tins.2008.03.006.
[authors' prepublication MSWord
document] [email for PDF: dgking@siu.edu]
This review emphasizes the applicability of our tuning-knob hypothesis (1997b, 1997c,
2006a) in the domain of neuroscience. My principal roles in the preparation of this article were recruiting
coauthors with expertise on the effects of simple sequence repeat variation, editing the tens of thousands of words provided
by these coauthors into the word-limit imposed by the journal, and writing the introduction and conclusion.
(2007c) King, D.G., What's so special about giant fibers? (slide show pdf)
This slide show was prepared for a symposium on myelin (cf.
evolution of myelin), at the National Evolutionary Synthesis Center
(the precursor to TriCEM).
Content is similar to 2013a (above). My invitation to this symposium came from Dan Hartline,
who had been a professor down the hall from Allen Selverston's laboratory at UC San Diego while I was a graduate student in that
lab.
(2007b) King, D.G., and Y.
Kashi Mutation rate variation in eukaryotes: evolutionary implications of
site-specific mechanisms. Nature Reviews Genetics 8 (November 2007)
| doi:10.1038/nrg2158-c1.
This is the second of two short essays published in 2007, in response to published papers which failed to address overwhelming
evidence for extensive variation in localized, site-specific mutation rates. Unfortunately, the significance of such
sites is still being ignored years later (e.g.,
Zhang 2022, also in Nature Reviews Genetics). Mutation rate continues to be treated as a single, unitary parameter, while
Sturtevant's obsolete 1937 dictum -- that "mutations are accidents, and accidents will happen" -- persists as misleading dogma (cf.,
Indirect selection of implicit mutation protocols, above).
(2007a) King, D.G., and Y.
Kashi Mutability and Evolvability: Indirect selection for mutability.
Heredity 99:123-124. doi:10.1038/sj.hdy.6800998.
[email for PDF: dgking@siu.edu]
This is the first of two short essays published in 2007, in response to published papers which failed to address overwhelming
evidence for extensive variation in localized, site-specific mutation rates. Unfortunately, the significance of such
sites is still being ignored years later (e.g.,
Zhang 2022). Mutation rate continues to be treated as a single, unitary parameter, while
Sturtevant's obsolete 1937 dictum -- that "mutations are accidents, and accidents will happen" -- persists as misleading dogma (cf.,
Indirect selection of implicit mutation protocols, above).
(2006e) King, D.G., Facilitating Evolution:
Simple Sequence Repeats Make Genes Adjustable. (poster pdf).
While attending a conference at UC Berkeley to present this poster, I shared a hotel room with John Fondon who had made
a splash in 2004 by reporting the role of repetitive DNA in morphological variation of dogs. I later recruited
Fondon as a coauthor for our Trends in Neuroscience paper (2008a).
(2006d) Y.
Kashi and D.G. King Has simple sequence repeat mutability been selected
to facilitate evolution? Israel Journal of Ecology and Evolution
52:331-342. [Abstract]
[email for PDF: dgking@siu.edu]
This essay continued our campaign to publicize the evolutionary implications of simple sequence repeats.
(2006c) Hammock, E.A.D., and D.G. King Genes
and neuroethology: How can evolution adjust behavior? International
Society for Neuroethology Newsletter, November 2006, pp. 4-7.
This newsletter article highlighted new information relating simple sequence repeats to affiliative behavior in voles, as
reported by Elizabeth Hammock in the previous year.
I later recruited Hammock as a coauthor for our Trends in Neuroscience paper (2008a).
(2006b) Y.
Kashi and D.G. King Simple
Sequence Repeats as Advantageous Mutators in Evolution. Trends in Genetics
22: 253-259. [Abstract]
doi: 10.1016/j.tig.2006.03.005
[email for PDF: dgking@siu.edu]
This review updated our 1997 TiG review, with greater emphasis on evolutionary implications.
It has become our most highly-cited publication.
☞
Evidence for such "advantageous mutators" has continued to accumulate. Some of the more recent publications are cited in my 2024 article
in The Journal of Physiology, which generalizes the concept to additional protocols for mutation besides tandem repeats.

(2006a) King, D.G., E.N.
Trifonov, and Y.
Kashi Tuning Knobs in the Genome: Evolution of Simple Sequence
Repeats by Indirect Selection. In: Lynn H. Caporale, ed.,
The
Implicit Genome, Oxford University Press. [Abstract]
[Table
of contents for The Implicit Genome] [email for preprint:
dgking@siu.edu]
Lynn Caporale offered the invitation to submit this essay, based on mutual interests established at her 1998 NYAS meeting, "Molecular strategies
of biological evolution" (1999b). In previous reviews coauthored with Kashi and Soller (listed below), I had depended
for content almost entirely on those two coauthors. Preparing to write this review finally gave me enough confidence to continue
writing on my own.
Another essay in this volume, "Epilogue: An Engineering Perspective: The Implicit Protocols,"
by John Doyle, Marie Csete, and Lynn Caporale, introduced the "protocols" metaphor which became central
to my 2012 essay in Annals NYAS.
(2002) King, D.G., and J.G. Killian Binocular rivalry, bipolar
disorder, and aging: Perceptual alternation rate correlates with age as
well as with psychiatric diagnosis. Program No. 803.4 Orlando FL: Society
for Neuroscience. [Abstract
with graph of data] [extended discussion]
Prompted by events in my personal life, I put aside my principal interest in evolutionary
genetics for a three-year project summarized in this abstract. The project was funded by the
Brain & Behavior Research Foundation
(which I had learned about through membership in NAMI). Its subject,
binocular rivalry,
is a curious phenomenon with implications for the neural substrate of consciousness
(more).
Killian, a graduate student at the time, gave up a secure job to assist with this project.
(2001) King, D.G. Evolution
of neural circuits. SIUC Cognitive Science Colloquium.
This text of a seminar presentation on October 5, 2001, summarizes most of the
central ideas of my research career, both before and after this date.
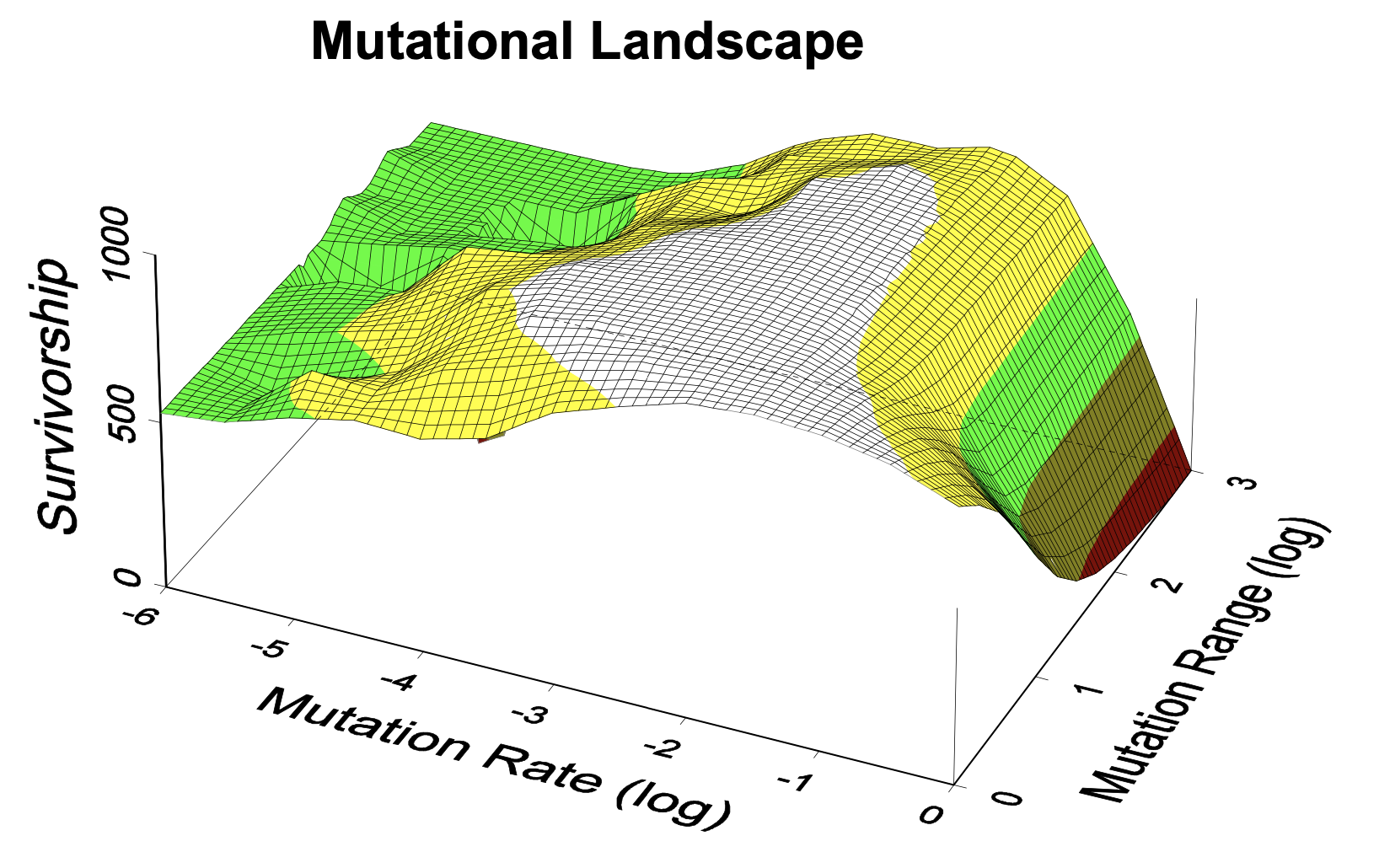
(2000) King, D.G. Indirect selection on the mutational landscape:
An evolved role for the mutability of repetitive DNA? Poster #218, in Evolution 2000,
(ed. M.J. Wade), Proceedings of the 2000 annual meeting of the Society for the Study of Evolution. [extended abstract, pdf]
An extended abstract of this meeting presentation, including graphically illustrated results of a simple model of population genetics,
was published in a CD-ROM of the proceedings of the 2000 meeting of the Society for the Study of Evolution. (Unfortunately, the full contents of this CD
do not appear to be available online.) Disappointingly, this poster presentation garnered essentially no interest; reaching a broad audience with
this idea has proven challenging (cf. 2007a and 2007b, above).
(1999c) Krajewski,
C., Fain, M., L. Buckley and King, D. G. (1999) Dynamically
heterogenous partitions and model-based phylogenetic inference: An evaluation
of analytical strategies with cytochrome b and ND6 gene sequences in cranes.
Molecular Phylogenetics and Evolution 13: 302-313.
[Abstract]
For this and a 1996 paper also coauthored with Krajewski, my only contribution was the coding of conceptually-simple computer programs
which facilitated the comparison of DNA sequences. Matt Fain and Larry Buckley were Krajewski's graduate students.
(1999b) King, D.G. Modelling selection for adjustable genes based
on simple sequence repeats. In: Molecular Strategies in Biological Evolution,
Annals of the New York Academy of Science 870: 396-399.
This abstract was published in the proceedings of a 1998 meeting of the New York Academy of Sciences, "Molecular strategies in
biological evolution," organized by Lynn Helena
Caporale. Prior to this meeting, I had written a computer program simulating a very simple model of population genetics.
Output from many runs of that program had convinced me that the speculative idea of "indirect selection" could plausibly occur as a real
phenomenon. A more elaborate version of this computer model was prepared for the 2000 meeting of the Society
for the Study of Evolution.
I had travelled to this meeting not knowing what to expect. At the time I still felt like a naive outsider in
the field of molecular evolution. But at this meeting I found that there existed a small community of biologists
who shared my growing interest in molecular mechanisms that could plausibly function as strategies for evolution.
This discovery felt like a turning point in my career.
I was pleasantly astonished when the first speaker (James A. Shapiro)
cited our recent Trends in Genetics review (1997b). I was astonished when a subsequent speaker,
Ed Trifonov, suggested that a plausible metaphor for the behavior
of simple sequence repeats might be that of "tuning knobs." When I approached him after his talk with a reprint of our
"Evolutionary Tuning Knobs" essay (1997c), his reaction was priceless. We later
collaborated on a 2006 essay for Caporale's volume, The Implicit Genome.
Delightfully (for me),
my own participation at this meeting was reported in a "News Focus" article for the journal Science:
"How the Genome Readies Itself for Evolution" (E. Pennisi,
Science, 21 Aug 1998, Vol 281: 1131-1134, DOI: 10.1126/science.281.5380.1131).
(Incidently, my flight from Memphis to New York to attend this meeting had memorably passed through a spectacular "cathedral"
of stormclouds, as our airliner wove closely among columns of thunderheads that stretched for miles into the distance. In hindsight
this was a remarkable omen for subsequent events in my career.)
 (1999a) King, D.G., and M. Soller Variation and fidelity: The evolution of simple sequence repeats
as functional elements in adjustable genes. In: S.P. Wasser, ed.,
Evolutionary Theory and Processes: Modern Perspectives, pp. 65-82. Kluwer
Academic Publishers, Dordrecht, The Netherlands. [Abstract]
[email for PDF: dgking@siu.edu]
(1999a) King, D.G., and M. Soller Variation and fidelity: The evolution of simple sequence repeats
as functional elements in adjustable genes. In: S.P. Wasser, ed.,
Evolutionary Theory and Processes: Modern Perspectives, pp. 65-82. Kluwer
Academic Publishers, Dordrecht, The Netherlands. [Abstract]
[email for PDF: dgking@siu.edu]
This essay, published in a festschrift dedicated to Eviatar Nevo, continued our
campaign to publicize the evolutionary implications of simple sequence repeats.
(1998) King, D.G. Cells "R" Us. Brain Awareness Week at SIUC.
This entry is the text of a public lecture offered during a program of public educational activities
sponsored by the Society for Neuroscience, and the
Dana Foundation. My goal in this presentation was to acquaint attendees with the basic idea that nerve
cells, as the working parts of the brain, are intimate participants in sensation and movement.
(1997d) King, D.G. Is there
a role for comparative genetics in neuroethology? International
Society for Neuroethology Newsletter, July 1997.
This newsletter article was effectively a preview of subsequent 2006c and 2013a articles in the same society newsletter.
(1997c) King, D.G., M. Soller, and Y.
Kashi Evolutionary tuning knobs. Endeavour
21: 36-40. [publisher pdf] [email for PDF: dgking@siu.edu]
This essay properly introduced "tuning knobs" as a metaphor for the functionality of repetitive DNA (first mentioned in abstract two years earlier,
in 1995). This same metaphor had been independently suggested by Ed
Trifonov, at a 1998 meeting of the New York Academy of Sciences ("Molecular strategies in biological evolution," organized by
Lynn Helena Caporale). Trifonov's surprised
response when I showed him a reprint of this article led eventually to our collaboration on an essay published in 2006a,
"Tuning knobs in the genome: Evolution of Simple Sequence Repeats by Indirect Selection." A few years later he invited me to
participate in a
conference at Gregor Mendel's abbey in Brno.
I was quite pleased when the "evolutionary tuning knob" concept was picked up in an article in Scientific American
(January, 1999, pp. 94-99), "DNA Microsatellites: Agents of Evolution?",
by Richard Moxon and Chris Wills.
☞ I had initially encountered the metaphor of "twiddling a knob on a concept"
in an essay by Douglas Hofstadter, "Variations on a theme as the crux of creativity."
[This essay first appeared in Scientific American in October, 1982; it was subsequently reprinted in Metamagical Themas: Questing for
the Essence of Mind and Pattern, Basic Books 1985.] Variations on a theme are commonplace in biology. At that time,
while I was developing the idea of metaptation, I had begun to wonder how a genome might quantitatively explore
variations on a theme. I learned of a plausible answer in a 1983 essay, "Updating the theory of mutation," by Drake, Glickman,
& Ripley (American Scientist 71:621-630) which described the peculiar mutational characteristics of repetitive DNA.
But it took another decade for evidence to appear that DNA could indeed have such a function, leading first to my
1994 letter to Science and eventually to this "Evolutionary Tuning Knobs" essay.
The "tuning knob" metaphor also features prominently in two essays published in 2024, in The Journal of Physiology
and in Trends in Genetics, and another in 2025 in Trends in Ecology and Evolution.
 |
Chezi and me
(with Gregor Mendel) |
(1997b) Y. Kashi,
D.G. King, and M. Soller
Simple sequence repeats as a source of quantitative genetic variation.
Trends in Genetics 13: 74-78. [Abstract]
[email for PDF: dgking@siu.edu]
This review, my first collaborative publication with Morris Soller (Moshe)
and his former student Yechezkel Kashi (Chezi), has become one
of our most-cited publications (exceeded only by its sequel, 2006b). It helped establish the idea that
simple sequence repeats could be significant sources of genetic variation.
This collaboration had been prompted by my 1994 letter to Science.
After noticing that letter, Moshe reached out to me from the Hebrew University in Jerusalem, proposing
that we work together to publicize our shared idea. (Soller, coincidently, was an old friend of
Ed Simon,
my undergraduate genetics professor at Purdue; see my epilog.) To facilitate our
long-distance correspondence, Moshe initiated me into the use of email. I finally visited Moshe
in person in 2007, at his family home in Chicago. I met Chezi three years later, at a conference (2010b) held at
Gregor
Mendel's abby in Brno, Czech Republic.
(1997a) King, D.G. Behavioral adjustments
[letter to the editor], American Scientist 85: 300-301.
This letter addressed an essay, "The evolution of arthropod nervous
systems," which had been published in the journal's previous issue (Osirio, Bacon & Whitington, Amer. Sci., 85:244-253,
May/June 1997). My letter posed a simple question: How can a limited genome support extraordinarily facile evolution of
instinctive behavior? It then proposed that the answer might involve repetitive DNA. Alongside this letter, the journal
published a response by the authors of the original
essay, expressing strongly-worded skepticism. Such skepticism was common at the time (and still is, unfortunately), but was addressed that same year
by our articles in Endeavour and Trends in Genetics (1997b, 1997c, above) as well as in several of our subsequent
publications (e.g., 2008a), culminating in my 2013a essay for the International
Society for Neuroethology. (For a related, extended discussion, see my 2001 online-only essay, "Evolution
of neural circuits.")
(1996) Krajewski, C. and D.G. King, Molecular divergence
and phylogeny: Rates and patterns of cytochrome b evolution in cranes. Molecular Biology and Evolution 13: 21-30.
[Abstract]
Carey Krajewski is a friend and colleague at SIUC Zoology. For this and a subsequent 1999c
paper also coauthored with Krajewski, my only contribution was the coding of conceptually simple-minded computer programs
which facilitated the comparison of DNA sequences. (As simple as these programs were, I have heard that they've been
used by other researchers, from The Smithsonian Institution in Washington DC to LaTrobe University in Australia.)
(1995) King, D.G., Triplet-repeat DNA: Unstable regulatory genes
could be evolutionary "tuning knobs." (abstract only)
This abstract for a Neuroscience meeting represents my first published mention of the "tuning knobs"
metaphor, published more substantially two years later (1997c).

(1994) King, D.G. Triple repeat DNA as a highly mutable regulatory
mechanism. Science 263: 595-596. [publisher pdf]
[email for PDF: dgking@siu.edu]
This letter-to-the-editor (appearing in print just in time for my 46th birthday!) was prompted by a
news article in Science ("The Puzzle
of the Triple Repeats," 4 June 1993), which had reported a peculiar quantitative relationship between severity of
symptoms in Huntington's disease and number of CAG repeats in the causative gene. This relationship corresponded
remarkably well with a speculation I had conceived a decade earlier, while developing the concept of "metaptation."
By staking my claim to the hypothesis that repetitive DNA might confer upon genes the property of evolutionary adjustability,
this letter officially marked the beginning of my re-invention as a theoretical evolutionary geneticist. I was over the moon
when, one week after this letter appeared, Science published a paper by
Gerber et al. which powerfully confirmed the plausibility of my hypothesis.
Publication of this letter led directly to collaboration with Yechezkel
Kashi and Morris Soller, of the Hebrew University in
Jerusalem, who had independently developed similar ideas. Together we have coauthored several papers listed above,
beginning in 1997 with Trends in Genetics and Endeavour (1997b and 1997c).
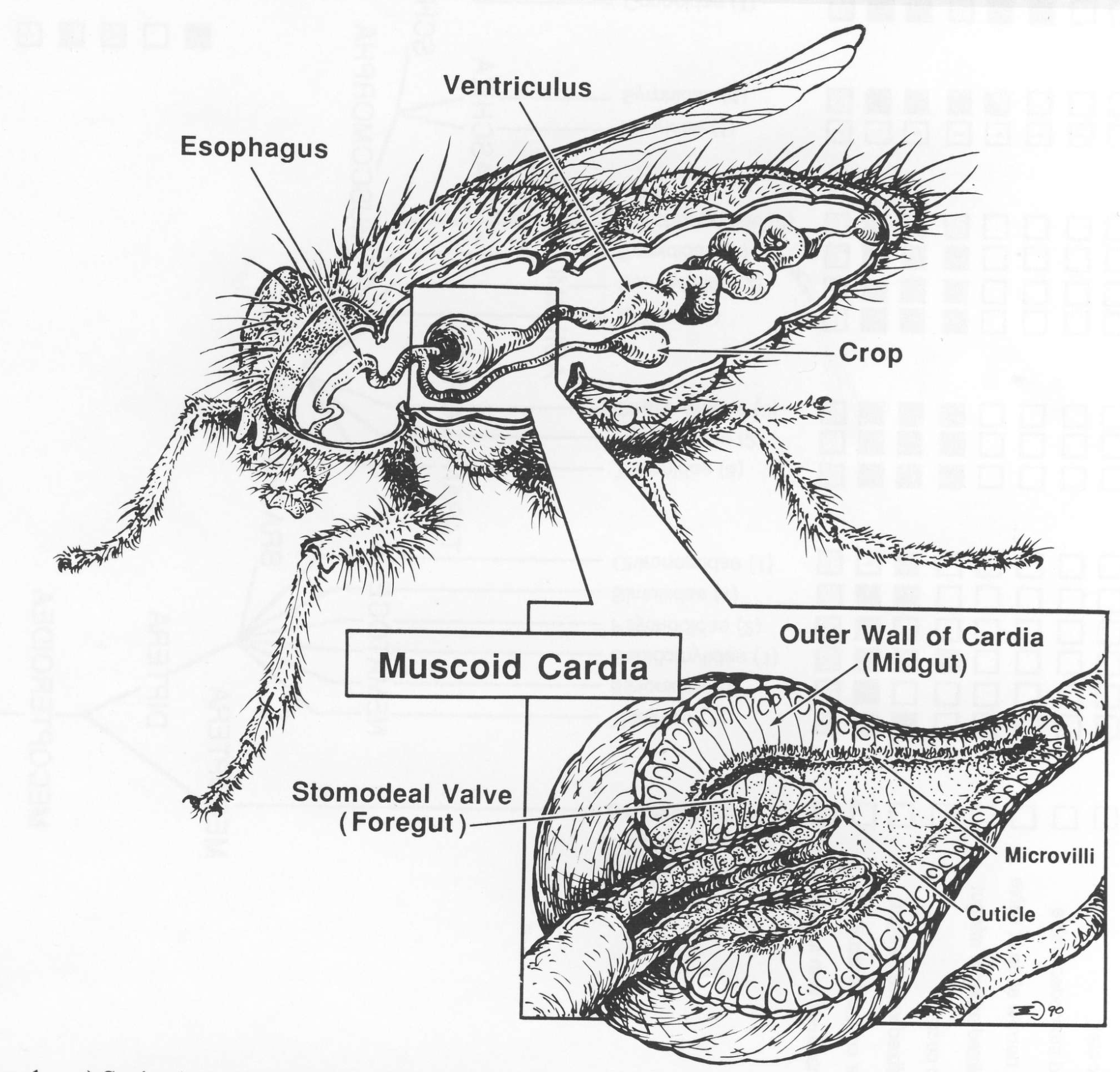
(1991) King, D.G. The origin of an organ: Phylogenetic
analysis of evolutionary innovation in the digestive tract of flies (Insecta:
Diptera). Evolution 45:568-588.
[email for PDF: dgking@siu.edu]
Research leading up to this report, which involved collecting and studying many diverse fly species, was probably the most fun of any
in my career. It provided intense engagement with evolutionary thinking and phylogenetic reasoning, and it resulted in an elegant story
of how a fairly complex organ had evolved. I became inordinately pleased with myself -- a neuroscientist by training -- for
successfully publishing in a journal devoted to evolutionary biology. (I had not yet begun my full transformation into an
evolutionary geneticist.) Although this report has not been cited nearly as often as several subsequent papers on molecular
evolution (e.g., 1997b, 1997c, 2006b), it remains one
of my favorite publications.
(1989) King, D.G. Phylogenetic diversity
of cellular organization in the cardia of muscoid flies (Diptera: Schizophora). Journal of Morphology
202: 435-455. [email for PDF: dgking@siu.edu]
This publication set the stage for my 1991 report on the evolutionary origin of the dipteran cardia. It was based on a collection
of various fly species initially intended for exploration of the phylogenetic origin of giant axons found in Drosophila, a study eventually published in
the ISN newsletter in 2013a.
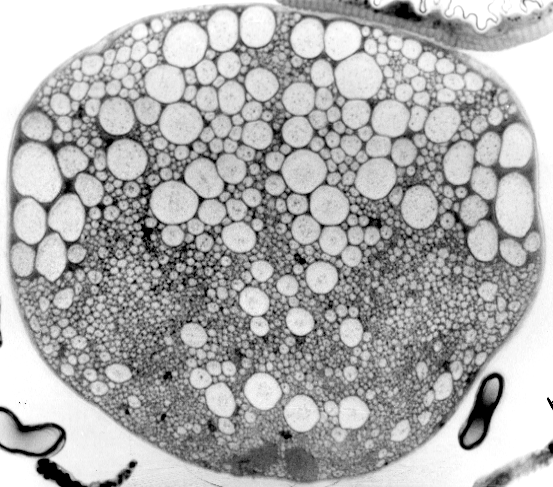
(1988b) King, D.G. On the evolution of specialized neuronal form: Phylogenetic variation in cervical
giant axons of flies (Diptera). Proc. XVIII Int. Congress
of Entomology, p. 70 (abstract only).
Travel to this Congress of Entomology meeting was especially memorable: My wife and I drove from southern Illinois to the meeting site
in Vancouver, B.C., touring the country for five weeks with our son and daughter, ages 6 and 9.
Several years earlier (in 1980), I had described a giant-axon pathway in Drosophila.
Shortly afterwards I had established that a homologous giant-axon pathway, although missing in tsetse flies
(1983a), could nevertheless be found in other muscoid flies (1983b).
In an effort to establish when this giant-axon pathway had first evolved, I set out to map
its phylogenetic distribution by examining as many different flies as I could easily acquire. This provided me
with a welcome excuse for outdoor fieldwork, collecting specimens around the delightful environs of southern Illinois.
Eventually, by the time of this meeting, I had recorded axon size distributions in sixty-five species representing
thirty-six families.
Although observations presented at this Vancouver meeting did not suggest a simple story
about the evolutionary origin of Drosophila giant fibers, they were nonetheless quite intriguing:
Axon size distributions among various fly species were unexpectedly diverse, even within a single family.
A few images of this diversity were eventually published 25 years later. And the same collection
of specimens did serve admirably for my analysis of the evolutionary origin of the dipteran cardia
(in 1989 and 1991).

(1988a) King, D.G. Cellular organization and peritrophic membrane formation
in the cardia (proventriculus) of Drosophila melanogaster. Journal
of Morphology 196: 253-282. [Abstract]
[email for PDF: dgking@siu.edu]
This study was prompted by accidental observation of abnormal cardia development in some behavioral mutants of Drosophila,
whereupon I realized that this organ had never received a detailed histological description. This was my first substantial
work that had not been grounded on work in the laboratories of my doctoral and post-doctoral advisors. It led directly to my 1989
and 1991 publications (above) on comparative anatomy of the Dipteran cardia.
I might never have paid attention to the fly cardia, but for its location atop the thoracic nervous system
where I could not help but notice it while studying nerve fibers.
(1985) King, D.G. Metaptation: A descriptive category for evolutionarily
versatile patterns of genetic and ontogenetic organization. Abstract only, in Evolutionary
Theory 7:222. [Abstract] [unpublished
ms.]
This abstract marked the beginning of a major shift in my career orientation, from neuroscience to evolutionary genetics, culminating
in a pre-retirement 2012 Annals NYAS paper and several post-retirement essays in 2024 and 2025,
all on mutation protocols as exemplified by short tandem repeats.
The central idea lying behind this abstract -- that effective evolution might require "adaptations for adaptability" -- emerged while
I was trying to understand what manner of genetic organization could possibly sculpt fine details of individually specialized nerve cells,
such as Drosophila's giant axons.
"Metaptation," the title neologism here, is modelled after Gould & Vrba's coinage of "exaptation."
This word has not yet entered mainstream usage, although it has been adopted by at least one researcher working with the evolvability of computer programs
(Moshe Looks 2007, Program evolution for general intelligence,
full text) and is elaborated
in William Calvin's book The River that Flows Uphill (1986, 2010).
Twenty-five years later, at a 2010b meeting in the Czech Republic, I presented a "new, improved" definition for
the word. And I took advantage of my post-retirement 2024 Journal of Physiology article
to slip the word "metaptation" into mainstream biology literature.
| Making variations on a theme is really the crux of creativity. |
| Douglas Hofstadter 1982
| |
This 1985 abstract was originally written to accompany a lengthy essay that had been written in 1983
and submitted to a journal where it languished for two years before being rejected. (Meanwhile I had moved
on with histological study of fly anatomy.) The original manuscript
included my first recognition of the plausibility of "evolutionary tuning knobs," an idea that was inspired
by a
1982 essay by Douglas Hofstadter but not published properly until 1997.
☞ Here I must acknowledge a special debt to
Steven Jay Gould, whose
monthly essays in Natural History magazine
inspired me to wonder about how evolution really works. Without the encouragement that Gould offered in his generous reply
to a "fan letter," in which I shared some initial thoughts about evolvability, I probably would have lacked confidence to continue
writing in a domain of biology where I had no formal training.
(1984) Wyman,
R.J., J.B. Thomas, L. Salkoff, and D.G. King The Drosophila giant fiber system. In: Neural Mechanisms of Startle
Behavior (R. Eaton, Ed.), Plenum Press, New York.
My inclusion as an author of this review, based on my participation as a postdoc in
Robert Wyman's lab at Yale (1976-1977, with publication in
(1980), was essentially honorary; I played no part in preparation of this book chapter for publication.
(1983e) King, D.G. Review of Genetic Neurobiology, by Hall, Greenspan, & Harris; in
Quarterly Review of Biology, 58:286.
When I wrote this very brief book review (less than 1/2 page), genetics was still a fairly new tool for studying nervous systems.
I have since come to believe that additional value can be obtained by using some of the more remarkable features of nervous systems
as inspiration for re-thinking certain aspects of genetics (cf. What can giant axons tell us about
genetics?, above).
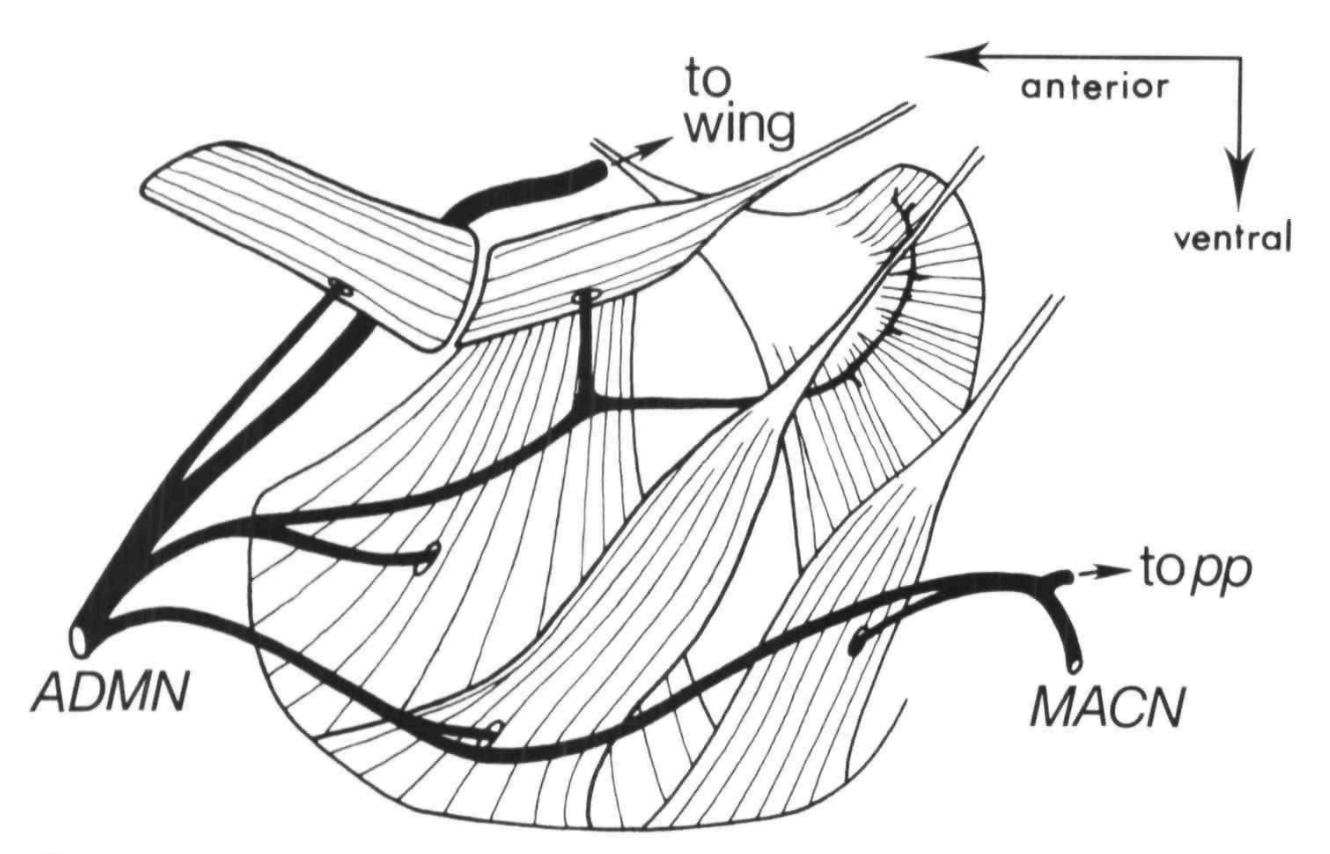
(1983d)
King, D.G. and M.A.
Tanouye Anatomy of motor axons to direct flight muscles in Drosophila. Journal
of Experimental Biology 105: 231-239.
While working as a postdoc in
Robert Wyman's lab at Yale, I had been delighted to discover that individual motor axons in a small animal the size of Drosophila
could be reliably traced through serial thin sections all the way from their neuromuscular junctions into the thoracic central nervous
system. This report of my histological observations was submitted jointly with Mark Tanouye, along with a companion paper
(1983c, below) reporting Tanouye's recordings of electrical activity from these muscles.
(1983c) Tanouye,
M.A. and D.G. King Giant fiber activation of direct flight muscles
in Drosophila. Journal of Experimental Biology 105: 241-251.
Two companion papers were submitted jointly with Mark Tanouye, a friend from our time together as postdocs in
Robert Wyman's lab at
Yale. This one, with Tanouye as principal author, reported physiological recording from Drosophila flight muscles.
My contribution to this paper was histological identification of sites where dye had been injected during recording. The other
paper of the pair (1983d, above) described the axons innervating those muscles.

(1983b) King, D.G. and K.L. Valentino On neuronal homology: A
comparison of similar axons in Musca, Sarcophaga and Drosophila (Diptera: Schizophora).
Journal of Comparative Neurology 219: 1-9.
[Abstract]
Establishing homology at the level of individual cells is fraught with both technical and conceptual difficulties. This article
discusses these challenges while reporting putative homology for a set of axons individually identified
in three fly species from three different taxonomic families. This was accomplished primarily by demonstrating that their similarity
extended to the level of synaptic location. Unpublished continuation of this work has demonstrated the same configuration of synapsing axons
in several additional fly species.
At the time this work was begun, Karen Valentino was an undergraduate student in
Robert Wyman's lab at Yale.
While working with Wyman as a postdoc, I offered her technical guidance as she prepared images of one of the specimens which
eventually appeared in this report.
(1983a) King, D.G. Evolutionary loss of a neural pathway from the
nervous system of a fly (Glossina morsitans, Diptera). Journal of Morphology
175: 27-32. [Abstract]
Together with my paper "On neuronal homology..." (1983b, above), this publication represented
my initial foray into comparative study and evolutionary thinking.
My interest in an evolutionary perspective had been kindled by
Steven Jay Gould's monthly essays in Natural History magazine, under the heading
"This view of life."
(1982b) King, D.G., N. Kammlade, and J. Murphy A simple device
to help reembed plastic sections. Stain Technology 57: 307-311. [Abstract]
Much of my histological study of flies involved relatively thick (2-5μm) serial sections
of plastic-embedded specimens. With some help from the gadget described in this report, selected sections could then be
lifted off their glass slides and re-sectioned for electron microscopy. (Ideally, as many as twenty sections
for electron microscopy might be cut parallel to the face of one 2μm thick section.)
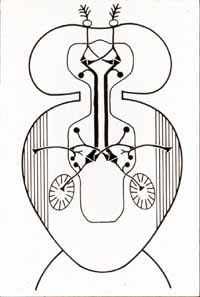
(1980) King, D.G. and R.J.
Wyman Anatomy of the giant fiber pathway in Drosophila: I. Three
thoracic components of the pathway. Journal of Neurocytology 9:
753-770. [Abstract]
This publication resulted from work completed during my post-doctoral experience (1976-1977) with
Robert Wyman at Yale, where my attention was redirected from examining identified nerve cells in lobster to investigating the unique giant axons
of Drosophila. The Roman numeral in the title betrays my optimistic but mistaken expectation that an additional publication would soon
follow.
Wyman's lab was focussed on the Drosophila giant fiber pathway in hopes of identifying genes responsible for such conspicuously
differentiated nerve cells. Imagining that phylogenic analysis might help illuminate the underlying genetics, I began looking
for giant axons in other fly species, beginning with Musca and Sarcophaga.
I subsequently collected images from 62 other species, a few of which were eventually published in a 2013a essay
entitled "What can giant axons tell us about genetics and evolution?"
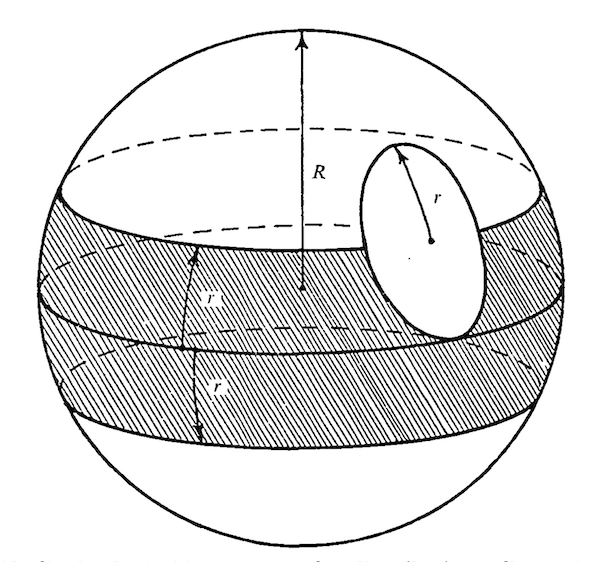
(1978) Wyman,
R.J. and D.G. King, The dispersion of imaginal disc progenitor cells
in the Drosophila blastoderm. Genetic Research 31: 273-286.
[Abstract]
My postdoctoral mentor, Robert Wyman,
was the principal author for this paper. My contribution -- a simple mathematical analysis
of a simplistic model of the formation of genetic mosaics in Drosophila -- included a hand-inked diagram (at right)
which inadvertently resembled the "death star" in the 1977 movie Star Wars.
(1976c) King, D.G. Organization of crustacean neuropil: II. Distribution
of synaptic contacts on identified motor neurons in lobster stomatogastric ganglion.
Journal of Neurocytology 5: 239-266. [Abstract]
[email for PDF: dgking@siu.edu]
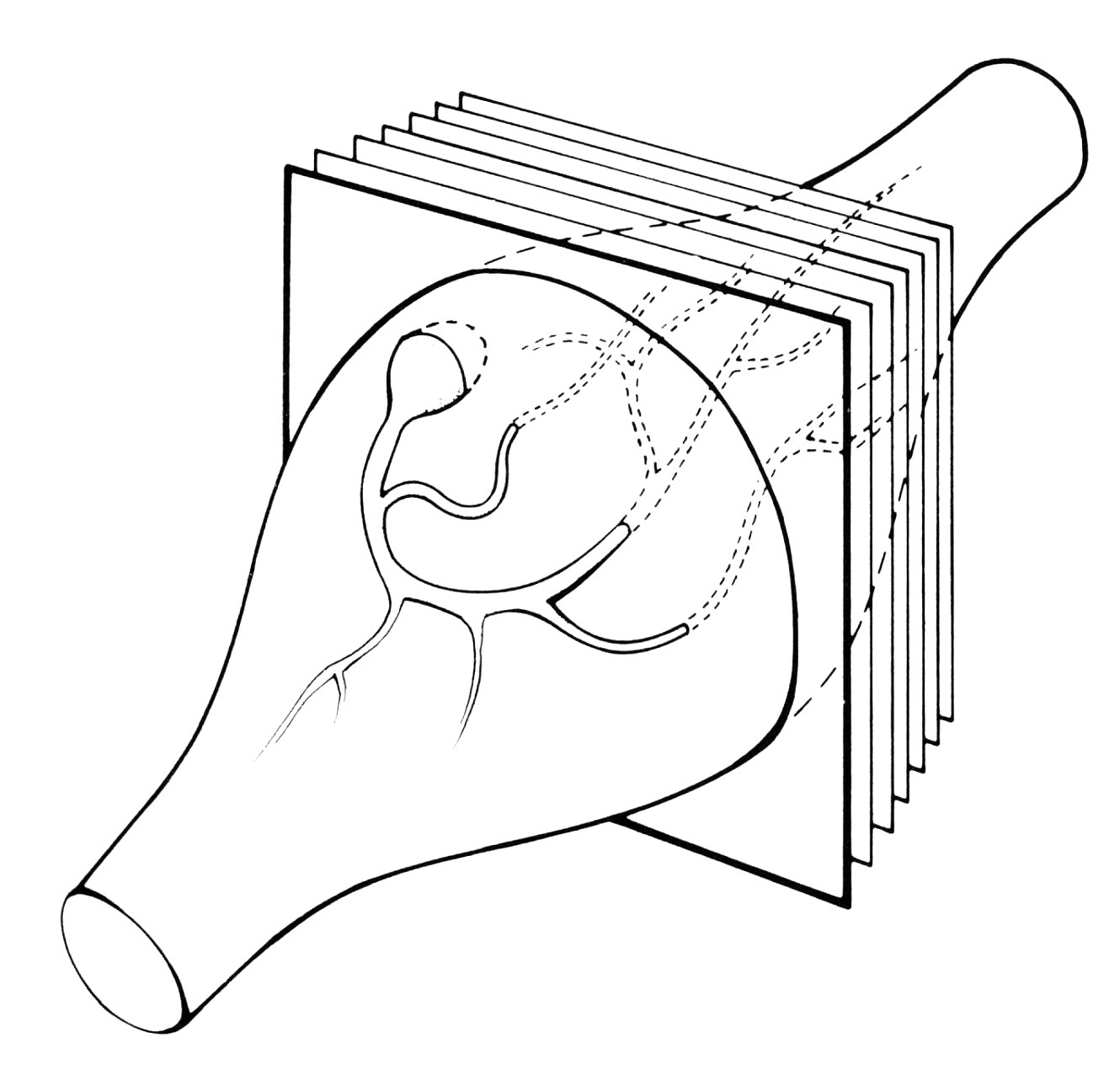
This paper, together with its companion paper (1976b, below), comprised my doctoral
dissertation in neuroscience at UCSD. (I was tickled pink when
this work was noticed in Scientific American [February 1978,
Gordon M. Shepherd, "Microcircuits in the Nervous System"],
a magazine I had read since childhood.)
This report is based on a single specimen whose cells had been individually identified electrophysiologically
by my dissertation advisor, Allen Selverston. Serial-sectioning
this specimen for electron microscopy enabled reconstruction
of the shape of each cell's principal branches, including localization of specific synapses. Cutting, staining, and imaging
approximately 5000 sections through this small piece of tissue (less than a millimeter in length), together with analyzing the resulting
micrographs, was by far the most strenuous single piece of lab-work in my career.
☞
The phrase "identified motor neurons" in this paper's title refers to uniquely identifiable
nerve cells. This concept -- still fairly new in the 1970s and remaining relatively unfamiliar even into the 21st
century -- became a guiding theme in my subsequent research. Without realizing it at the time, I was both lucky and
privileged to have been accepted as a student into an institution where "one of the major revolutions
in neuroscience" was underway!
(1976b) King, D.G. Organization of crustacean neuropil: I. Patterns
of synaptic connections in lobster stomatogastric ganglion. Journal
of Neurocytology 5: 207-237. [Abstract]
[email for full-text PDF: dgking@siu.edu]
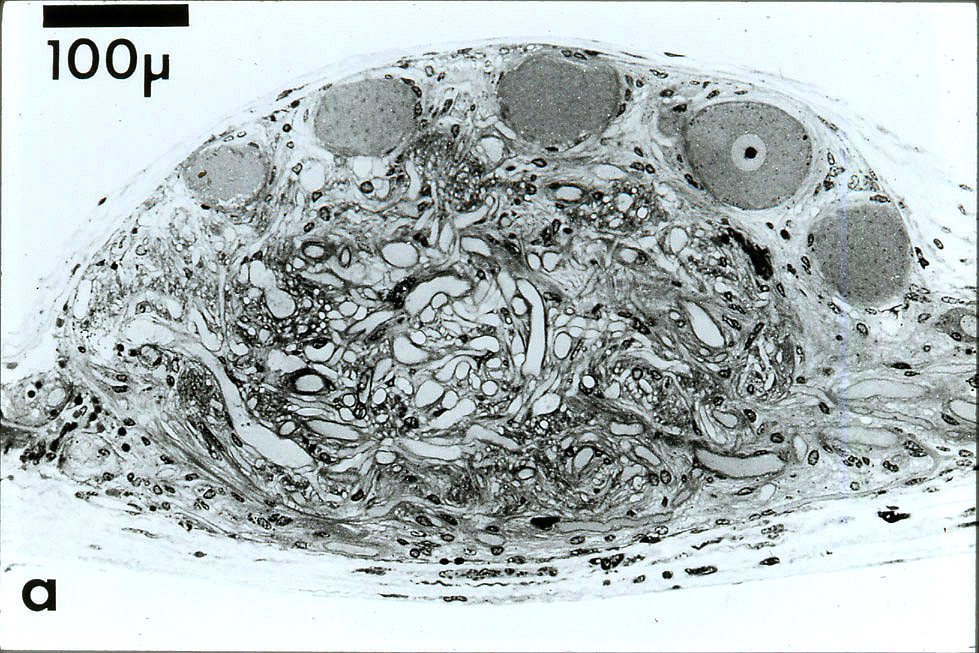 Remarkably, this paper is
still being cited 50 years after publication. Together with its companion paper (1976c, above), this paper comprised my doctoral
dissertation in neuroscience at the University of California San Diego, completed with
extensive support through the laboratory of Allen Selverston.
Remarkably, this paper is
still being cited 50 years after publication. Together with its companion paper (1976c, above), this paper comprised my doctoral
dissertation in neuroscience at the University of California San Diego, completed with
extensive support through the laboratory of Allen Selverston.
(1976a) Selverston,
A., D.R. Russell, J.P. Miller, and D.G. King, The stomatogastric
nervous system: Structure and function of a small neural network.
Progress
in Neurobiology 7: 215-290.
For several decades, a number of laboratories around the world have been intensively investigating the crustacean
stomatogastric nervous system -- a model system
for analyzing the mechanisms of motor pattern generation. I completed my own graduate study at one center for
such study, Allen Selverston's lab at UC San Diego. This publication is a review of work up to 1975.
[More recent reviews appeared in 1987,
and 2016.] My principal contribution (in addition to assisting with overall
editing) was a single-chapter preview of my doctoral dissertation, published separately and in greater detail in the two
1976 publications listed immediately above. (Coauthors Russell and Miller were also graduate students in Selverston's lab.)
until 1980 | 1981-1990 |
1991-2000 | 2001-2010 |
2011-2020 | since 2020 | Epilogue
Return to King homepage
 Comments and questions: dgking@siu.edu
Comments and questions: dgking@siu.edu
SIUC / Zoology / David King
https://dgkinglab.siu.edu/PUBLIST.HTM
Last updated: 20 February 2026 / dgk






















 Remarkably, this paper is
still being cited 50 years after publication. Together with its companion paper (1976c, above), this paper comprised my doctoral
dissertation in neuroscience at the University of California San Diego, completed with
extensive support through the laboratory of Allen Selverston.
Remarkably, this paper is
still being cited 50 years after publication. Together with its companion paper (1976c, above), this paper comprised my doctoral
dissertation in neuroscience at the University of California San Diego, completed with
extensive support through the laboratory of Allen Selverston.  Comments and questions:
Comments and questions: